Kim-Hellmuth Lab
Functional Immunogenomics
-


Research topics
- Genetic influence on human immune response variation
- Genetic ancestry effects on the human immune system
- Human perinatal immune cell development
- Molecular characterization of COVID-19 infections in children and young adults
Prof. Dr. med. Sarah Kim-Hellmuth
✉ Sarah.Kimhellmuth@med.uni-muenchen.de
Bluesky/🐦/X: @skimhellmuth
Profile at Die Junge Akademie: Click here
Profile at #InnovativeFrauen: Click here
-
Genetic basis of human immune response variation

The human immune system plays an important role in host protection, autoimmune and inflammatory diseases, cancer, metabolism, and ageing. Given this central role in many human pathologies, it is crucial to understand the variability of immune responses at the population level and how this variability relates to disease susceptibility.
Large genome-wide association studies have implicated hundreds of genetic loci in immune-related genes highlighting the immune system’s role in the biological mechanism underlying genetic risk to numerous diseases. However, for the vast majority of these genetic variants, we have little understanding of their functional effects and their context-specificity. Studying the genetic influence on immune response is further complicated by the complexity of the immune system, which consists of many different cell types that respond to a plethora of signals, interact with each other and induce different effector functions under diverse kinetics.
Our group integrates state-of-the-art genomic and functional genetic approaches to characterize the genetic basis of human immune response variation to advance our understanding of disease-associated variants and answer questions of genome function plasticity that is shaped by gene-by-environment interactions. We focus in particular on molecular quantitative traits (molQTLs) in the context of immune activation and disentangle the cell type and context specificity of functional genetic variants with the ultimate goal to develop a roadmap for complex traits at large and enable the move from genetic discovery to functional interpretation and ultimately clinical impact.
Genetic regulation of protein expression during immune cell development in human neonates
The composition and maturity of immune cells exhibits high interindividual variability in preterm and term human newborns. Quantitative and qualitative changes of neonatal neutrophils are thought to contribute to the increased susceptibility to bacterial infections and neonatal sepsis that are still a leading causes of mortality and long-lasting morbidity. This interindividual variability is shaped by genetically hard-wired programs, the rapidly changing environment, and the interaction between those two (GxE interaction). While elaborate perinatal models in mice and other model systems enable comprehensive research on environmental factors that influence perinatal immune cell development, the genetic basis of human perinatal immune cell variability is still poorly understood and largely understudied.
In this project we investigate the role of cis-regulatory variants on protein expression during perinatal immune cell development in human neonates. To do this, we use the Munich Preterm and Term clinical (MUNICH-PreTCl) birth cohort. Integrating proteome profiles and genotype information of each participant, we will identify genetic variants that determine the variability in protein abundance in neonatal blood samples at defined gestational time points. These so-called protein quantitative trait loci (pQTL) offer insight into how genetics shape individual phenotypes by understanding the subsequent modulation of distinct protein expression profiles.
COVID-19 in children and young adultsCOVID-19 is an infectious disease caused by the new strain of coronavirus SARS-CoV-2. It was first identified in 2019 in Wuhan, China, and has since spread globally, resulting in the 2019–20 coronavirus pandemic. Common symptoms include fever and cough, however disease symptoms as well as disease course and outcome are highly variable ranging from asymptomatic cases to severe pneumonia and death. While children are likely to have milder symptoms than adults, children of all ages are susceptible to COVID-19 and can suffer from severe disease. Until now it is unknown why children show a different course of disease compared to adults.
Studying the immune response to SARS-CoV-2 in children is therefore critical to rapidly advance our understanding of the pathophysiology of COVID-19 both in children and adults. Children offer a unique possibility to study host-related factors that determine COVID-19 severity in the absence of ageing and comorbidity-related interactions, which are largely determining the disease course in adults.
We have therefore initiated a functional genetics and genomics COVID-19 study to examine the genetic and environmental risk factors of COVID-19 in pediatric and adult patients. Our group integrates deep immune profiling with multi-omics across multiple molecular levels (genome, transcriptome, proteome, metabolome) to enhance our understanding of the human immune response to SARS-CoV-2. Following questions will be addressed:
- Why does SARS-CoV-2 affect children differently compared to adults?
- What are the genetic and immunological risk factors that contribute to this difference?
- Can we use these factors to identify those children who will become severely affected?
As part of the Child Health Alliance Munich (CHANCE) initiative this prospective study is performed at the Dr. von Hauner Children’s Hospital of the Ludwig-Maximilians University (LMU) Munich and the Department of Pediatrics of the Technical University of Munich School of Medicine (TUM). The study is also actively involved in national (Deutsche COVID-19 OMICS Initiative) and international (COVID-19 Host Genetics Initiative) COVID-19 initiatives to join forces in combating this pandemic.
-






















 Alumni
Alumni



-
Selected publications:
1. Puzek, B., Wratil, P. R., Janke, C., Le Thi, T. G., Rubio-Acero, R., Lupoli, G., Schoof, I. C., Stern, M., Zhelyazkova, A., Rech, J., Choukér, A., Koletzko, B., Wieser, A., Török, H. P., Castelletti, N., Keppler, O. T., Geldmacher, C., Hornung, V., Zeggini, E., Adorjan, K., Hoelscher, M., Koletzko, S., & Kim-Hellmuth, S. (2025). Genetic variation in HLA, IGKV and HHEX loci influence mRNA vaccine-induced long-lasting humoral protection against COVID-19. medRxiv, 2025.10.21.25337971. https://doi.org/10.1101/2025.10.21.25337971
2. Franz, T. M., Punathil, R. K., Soto-Beasley, A. I., Strongosky, A., Walton, R. L., Kim-Hellmuth, S., Springer, W., Dulski, J., Ross, O. A., Jaramillo-Koupermann, G., Alarcon, F., & Wszolek, Z. K. (2025). Screening for PRKN and PINK1 mutations in Ecuadorian patients with early-onset Parkinson's Disease. Neurologia i neurochirurgia polska, 59(1), 56–61.
3. Santos-Peral, A., Zaucha, M., Nikolova, E., Yaman, E., Puzek, B., Winheim, E., Goresch, S., Scheck, M. K., Lehmann, L., Dahlstroem, F., Karimzadeh, H., Thorn-Seshold, J., Jia, S., Luppa, F., Pritsch, M., Butt, J., Metz-Zumaran, C., Barba-Spaeth, G., Endres, S., Kim-Hellmuth, S., Waterboer, T., Krug, A. B., & Rothenfusser, S. Basal T cell activation predicts yellow fever vaccine response independently of cytomegalovirus infection and sex-related immune variations. Cell Rep Med, 6(2), 101946 (2025).
4. Schmidt, A., Casadei, N., Brand, F., Demidov, G., Vojgani, E., Abolhassani, A., Aldisi, R., Butler-Laporte, G., DeCOI host genetics group, Alawathurage, T. M., Augustin, M., Bals, R., Bellinghausen, C., Berger, M. M., Bitzer, M., Bode, C., Boos, J., Brenner, T., Cornely, O. A., Eggermann, T., Erber, J., Feldt ,T., Fuchsberger, C., Gagneur, J., Göpel, S., Haack, T., Häberle, H., Hanses, F., Heggemann, J., Hehr, U., Hellmuth, J. C., Herr, C., Hinney, A., Hoffmann, P., Illig, T., Jensen, B. O., Keitel, V., Kim-Hellmuth, S., [...], Motameny, S., Nothnagel, M., Riess, O., Schulte, E. C., & Ludwig, K. U. Systematic assessment of COVID-19 host genetics using whole genome sequencing data. PLoS Pathog 20(12) (2024).
5. Nussbaum, C. & Kim-Hellmuth, S. Unlocking the genetic influence on milk variation and its potential implication for infant health. Cell Genom. 4, 100676 (2024).
6. Kasela, S., Aguet, F., Kim-Hellmuth, S., Brown, B. C., Nachun, D. C., Tracy, R. P., Durda, P., Liu, Y., Taylor, K. D., Johnson, W. C., Berg, D. V. D., Gabriel, S., Gupta, N., Smith, J. D., Blackwell, T. W., Rotter, J. I., Ardlie, K. G., Manichaikul, A., Rich, S. E., Barr, R. G. & Lappalainen, T. Interaction molecular QTL mapping discovers cellular and environmental modifiers of genetic regulatory effects. Am J Hum Genet 111(1) (2024).
7. Kerimov, N., Tambets, R., Hayhurst, J. D., Rahu, I., Kolberg, P., Raudvere, U., Kuzmin, I., Chowdhary, A., Vija, A., Teras, H. J., Kanai, M., Ulirsch, J., Ryten, M., Hardy, J., Guelfi, S., Trabzuni, D., Kim-Hellmuth, S., Rayner, W., Finucane, H., Peterson, H., Mosaku, A., Parkinson, H. & Alasoo, K. eQTL Catalogue 2023: New datasets, X chromosome QTLs, and improved detection and visualisation of transcript-level QTLs. PLoS Genet. 19, e1010932 (2023).
8. Flynn, E., Tsu, A., Kasela, S., Kim-Hellmuth, S., Aguet, F., Ardlie, K. G., Bussemaker, H. J., Mohammadi, P. & Lappalainen, T. Transcription factor regulation of eQTL activity across individuals and tissues. Plos Genet 18 (2022).
9. Brandt, M. K., Kim-Hellmuth, S., Ziosi, M., Gokden, A., Wolman, A., Lam, N., Recinos, Y., Hornung, V. K., Schumacher, J. & Lappalainen, T. An autoimmune disease risk variant: A trans master regulatory effect mediated by IRF1 under immune stimulation? PLoS Genet. 17(7), (2021).
10. Warnat-Herresthal, S., Schultze, H., Shastry, K. L., Manamohan, S., Mukherjee, S., Garg, V., Sarveswara, R., Händler, K., Pickkers, P., Aziz, N. A., Ktena, S., Tran, F., Bitzer, M., Ossowski, S., Casadei, N., Herr, C., Petersheim, D., Behrends, U., Kern, F., Fehlmann, T., Schommers, P., Lehmann, C., Augustin, M., Rybniker, J., Altmüller, J., Mishra, N., Bernardes, J. P., Krämer, B., Bonaguro, L., Schulte-Schrepping, J., Domenico, E. D., Siever, C., Kraut, M., Desai, M., Monnet, B., Saridaki, M., Siegel, C. M., Drews, A., Nuesch-Germano, M., Theis, H., Heyckendorf, J., Schreiber, S., Kim-Hellmuth, S., […], Deutsche COVID-19 Omics Initiative (DeCOI), Giamarellos-Bourboulis, E. J., Kox, M., Becker, M., Cheran, S., Woodacre, M. S., Goh, E. L. & Schultze, J. L. Swarm Learning for decentralized and confidential clinical machine learning. Nature 1–7 (2021).
11. de Goede, O. M., Nachun, D. C., Ferraro, N. M., Gloudemans, M. J., Rao, A. S., Smail, C., Eulalio, T. Y., Aguet, F., Ng, B., Xu, J., Barbeira, A. N., Castel, S. E., Kim-Hellmuth, S., Park, Y., Scott, A. J., Strober, B. J., GTEx Consortium, Brown, C. D., Wen, X., Hall, I. M., Battle, A., Lappalainen, T., Im, H. K., Ardlie, K. G., Mostafavi, S., Quertermous, T., Kirkegaard, K. & Montgomery, S. B. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell (2021).
12. GTEx Consortium#. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318-1330 (2020). #Lead analyst: Kim-Hellmuth, S.
>> Science cover and Introduction to Special Issue
>> Press releases: here and here
13. Kim-Hellmuth, S.†*, Aguet, F.*, Oliva, M., Muñoz-Aguirre, M., Kasela, S., Wucher, V., Castel, S. E., Hamel, A. R., Viñuela, A., Roberts, A. L., Mangul, S., Wen, X., Wang, G., Barbeira, A. N., Garrido-Martin, D., Nadel, B., Zou, Y., Bonazzola, R., Quan, J., Brown, A., Martinez-Perez, A., Soria, J. M., GTEx Consortium, Getz, G., Dermitzakis, E., Small, K. S., Stephens, M., Xi, H. S., Im, H. K., Guigó, R., Segre, A. V., Stranger, B. E., Ardlie, K. G. & Lappalainen, T. Cell type specific genetic regulation of gene expression across human tissues. Science 369 (2020).
>> Featured in 'This week in Science'
>> Press releases: here and here
14. Oliva, M.*, Muñoz-Aguirre, M.*, Kim-Hellmuth, S.*, Wucher, V., Gewirtz, A., Cotter, D., Parsana, P., Kasela, S., Balliu, B., Viñuela, A., Castel, S. E., Mohammadi, P., Aguet, F., Zou, Y., Khramtsova, E., Skol, A., Garrido-Martin, D., Reverter, F., Brown, A., Evans, P., Gamazon, E., Payne, A., Bonazzola, R., Barbeira, A. N., Hamel, A. R., Martinez-Perez, A., Soria, J. M., GTEx Consortium, Pierce, B., Stephens, M., Eskin, E., Dermitzakis, E., Segre, A. V., Im, H. K., Engelhardt, B., Ardlie, K. G., Montegomery, S., Battle, A., Lappalainen, T., Guigó, R. & Stranger, B. E. The impact of sex on gene expression and its genetic regulation across human tissues. Science 369 (2020).
>> Science Perspective and selected media coverage: Deutschlandfunk Forschung aktuell
>> Press releases: here and here
15. Demanelis, K., Jasmine, F., Chen, L. S., Chernoff, M., Tong, L., Delgado, D., Zhang, C., Shinkle, J., Sabarinathan, M., Lin, H., Ramirez, E., Oliva, M., Kim-Hellmuth, S., Stranger, B. E., Lai, T.-P., Aviv, A., Ardlie, K. G., Aguet, F., Ahsan, H., GTEx Consortium, Doherty, J. A., Kibriya, M. G. & Pierce, B. L. Determinants of telomere length across human tissues. Science 369 (2020).
16. Kim-Hellmuth, S.†, Bechheim, M., Pütz, B., Mohammadi, P., Nédélec, Y., Giangreco, N., Becker, J., Kaiser, V., Fricker, N., Beier, E., Boor, P., Castel, S. E., Nöthen, M. M., Barreiro, L. B., Pickrell, J. K., Müller-Myhsok, B., Lappalainen, T., Schumacher, J. & Hornung, V. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun 8, 266 (2017).
17. Kim-Hellmuth, S. & Lappalainen, T. Concerted Genetic Function in Blood Traits. Cell 167, 1167–1169 (2016).
18. Hornung, V., Ellegast, J., Kim, S., Brzózka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K.-K., Schlee, M., Endres, S. & Hartmann, G. 5'-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006).
†Corresponding author, *Equally contributing author
-
11/2025

Paula Rothämel at the CoPILOT Retreat

Pauline Kuschel at the CoPILOT Retreat

Maria Corey at the CoPILOT Retreat
10/2025
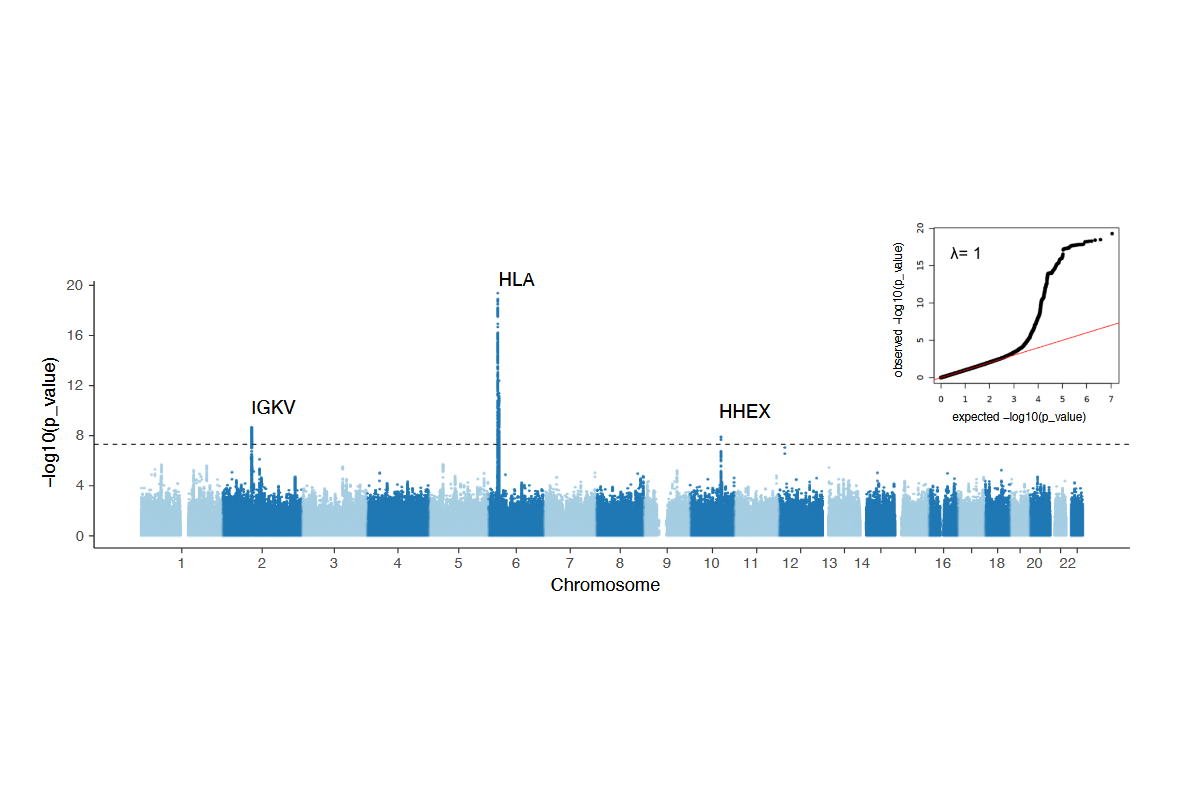
Our preprint on the influence of genetic variation on long lasting COVID-19 vaccine immunity
09/2025

MD student Francesco Zocchi joins the lab

Medical student Eva Herold joins the lab
08/2025

Medical student Maria Corey joins the lab
07/2025
Sathya Darmalinggam defends her PhD thesis
06/2025

04/2025

Medical student Fintan Viebahn joins the lab
03/2025

PhD student Irene Namara joins the lab


01-02/2025



Jessica Jin presents at the DGfI AKDC group meeting
202409/2024

Senior Bioinformatician Susanne Horn, PhD joins the lab

Medical student Chiara Geiger joins the lab

PhD student Furkan Büyükgöl joins the lab
07/2024

Clinician scientist Dr. med. Paula Rothämel joins the lab
06/2024

04/2024

Staff scientist Juliane Klein joins the lab

Clinician scientist Dr. med. Jessica Jin joins the lab
03/2024

Official start of our ImmGenDC Uganda team in Kyamulibwa

Master student Franziska Schweiger joins the lab

Master student Deniz Eser joins the lab

Medical student Sophia Grotz joins the lab
02/2024

01/2024

Kick-off meeting of our ERC Starting Grant project ImmGenDC with our collaborators at MRC Uganda

Postdoc Marie Bourdon, PhD joins the lab

MD student Aleksandra Jucha joins the lab
202312/2023

Medical student Mara Franzl joins the lab
11/2023

10/2023

Panel discussion at the Retreat of the LMU Medical & Clinician Scientist Program (MCSP)

PhD student Alessandro Mattia joins the lab
09/2023

Sathya Darmalinggam's poster presentation at the first joint conference of the French and German Societies of Immunology

Tuguldur Tumurbaatar's poster presentation at the first joint conference of the French and German Societies of Immunology
06/2023

Medical student Laura Jänisch joins the lab
05/2023

Sarah Kim-Hellmuth is portrayed in the children’s book “Young Scientists” of Die Junge Akademie
04/2023

MD student Mirjam Schobel joins the lab

Master student Pauline Kuschel joins the lab
03/2023
Bioinformatician Nicolás Lichilín, PhD joins the lab
Medical student Lea Steinberg joins the lab
02/2023


Research fellow Dr. med. Tobias Franz joins the lab
01/2023

Master student Preeti Thiyagarajan joins the lab

Medical student Ekin Yaman joins the lab
202211/2022


Master student Tuguldur Tumurbaatar joins the lab

Medical student Jana Wiesel joins the lab
09-10/2022
07-08/2022


Master student Nishant Chintalagiri joins the lab
05-06/2022

Panel discussion at the state occasion for the 175th birthday of the Hauner Children's hospital

03-04/2022

MD student Theresa Haslbeck joins the lab
202111/2021

PhD student Barbara Puzek joins the lab

10/2021
09/2021

PhD student Luise Zeckey joins the lab
05-06/2021


04/2021


Official start of the Helmholtz Young Investigator Group "Immunogenomics"
03/2021
 2020
202011-12/2020


MD student Alina Czwienzek joins the lab
10/2020

PhD student Sathya Darmalinggam joins the lab

Medical student Jöran Sarazzin joins the lab
09/2020


08/2020

Medical student Anda Ardeoan joins the lab
06/2020

04/2020

MD student Carola Kaltenhauser joins the lab


03/2020

The Ped-COVID-19 study is launched

-

Lab outing Summer 2025
Congratulations on your PhD, Sathya!

Our group at Helmholtz Girls' Day 2025

Poster presentation at the DGfI AKDC group meeting

Poster presentation at the CoPILOT retreat
ImmGenDC Uganda team meeting

Attending the CSAMA summer school in Brixen, Italy
Marie and Jessica at the Wellcome Genome Campus

Cake day after ESHG 2024

Our group at ESHG 2024
Visit to Oide Wiesn

Our group at the SFI-DGfI 2023 joint conference

ITG Retreat 2023 at Starnberger See

Lab outing Summer 2023

Welcome lunch with our new members Jana & Tuulu

First CompHealth retreat at HMGU

Sathya and Barbara at CGSI 2022

Our group at the Hauner concert for children in Ukraine
Lab outing Summer 2022
Welcome lunch with our new MD student Theresa

Lab after our first Ped-COVID-19 symposium

Welcome lunch with our new PhD student Luise

First lab outing after a long lockdown
-

Open positions:
We are actively looking to expand our group at all levels (postdocs, senior and junior staff scientists, graduate students, interns). Given our interdisciplinary work we offer both dry and wet lab projects and are looking for highly motivated individuals with diverse scientific backgrounds (immunologists, clinicians, computational biologists, statistical geneticists, data scientists).
- Fascinated by the immune system and its complexity and diversity among us human beings?
- Driven to better understand genetic gene regulation and its role in immune-related diseases?
- Interested in applying cutting-edge genomics technologies and analytical approaches to answer some of the pressing questions in genetics & genomics and immunology?
- Enjoy working in an interdisciplinary and collaborative team surrounded by a vibrant scientific environment?
Then look no more!
Our group is dedicated to better understand the genetic basis of human immune response variation and translate those insights into the clinic to improve every day medical decision-making. Located both at Helmholtz Munich and the LMU University hospital we are embedded and collaborate in a wide scientific network, in particular with the Computational Health Center, the Department of Pediatrics, the Biomedical Center, the Gene Center Munich and of course nationally and internationally.
If you are interested in discussing projects or job opportunities please get in touch via email. This can be even very early before your potential start or before major conferences to meet informally.
Diversity and inclusivity are key values in our group. We welcome applicants from all backgrounds especially from underrepresented and underprivileged backgrounds.
-
Kim-Hellmuth Lab
Dr. von Hauner Children's Hospital, University Hospital LMU Munich
Lindwurmstraße 4+49-89-4400-519821 +49-89-4400-57418
80337 MunichRgpgzeÜlvziäävfbzvim/fulrvfiuyziu/mi